Mutation = Phylogenetic Evolution.
Not content with inventing the most recent ‘killer mutation of death’, there are now apparently even more new strains of the ‘killer virus of death’ (the one that continues to not kill people, especially those it infects). There is now even one supposedly from South Africa (“this is an illegal mutating, you have two minutes to disperse”), which at least proves something can get into this country and isn’t stuck at Dover.
So, do you honestly think that these are new, killer mutant strains of death or are they just the results of antigen drift and to a lesser-degree antigen shift, a process that is natural in all viruses?
For starters, for ‘mutate’ think ‘degrade’ as mutation in the context of viral transcription & replication means a loss of quality control and a resultant ‘sub-standard’ output. By their very nature, RNA viruses mutate at a greater rate than DNA viruses because of higher levels of instability in their RNA dependent RNA polymerase [Sanjuán et al, 2010].
The presence of the exoribonuclease within nonstructural protein 14 (nsp14-ExoN) of SARS-CoV-2’s genome makes mutation far less likely, as nsp14-ExoN acts like a proofreader to maintain high fidelity replication. Think high quality photocopier. Therefore, in the first, instant, you can ignore everything that talks about mutations being a) a bad thing and b) unique to SARS-CoV-2.
Over time, viruses evolve. That’s phylogenetic evolution and is the reason why alphacoronaviruses and betacoronaviruses diverged over 7,150 years ago. So much for ‘new’.
Antigen Shift In The Spike Protein.
What is happening is antigen shift around the structural spike glycoprotein through glycosylation, something we first set out in October. The dynamic shifting of the glycan shielding is the virion attempting to protect its antigen through immunoevasion.
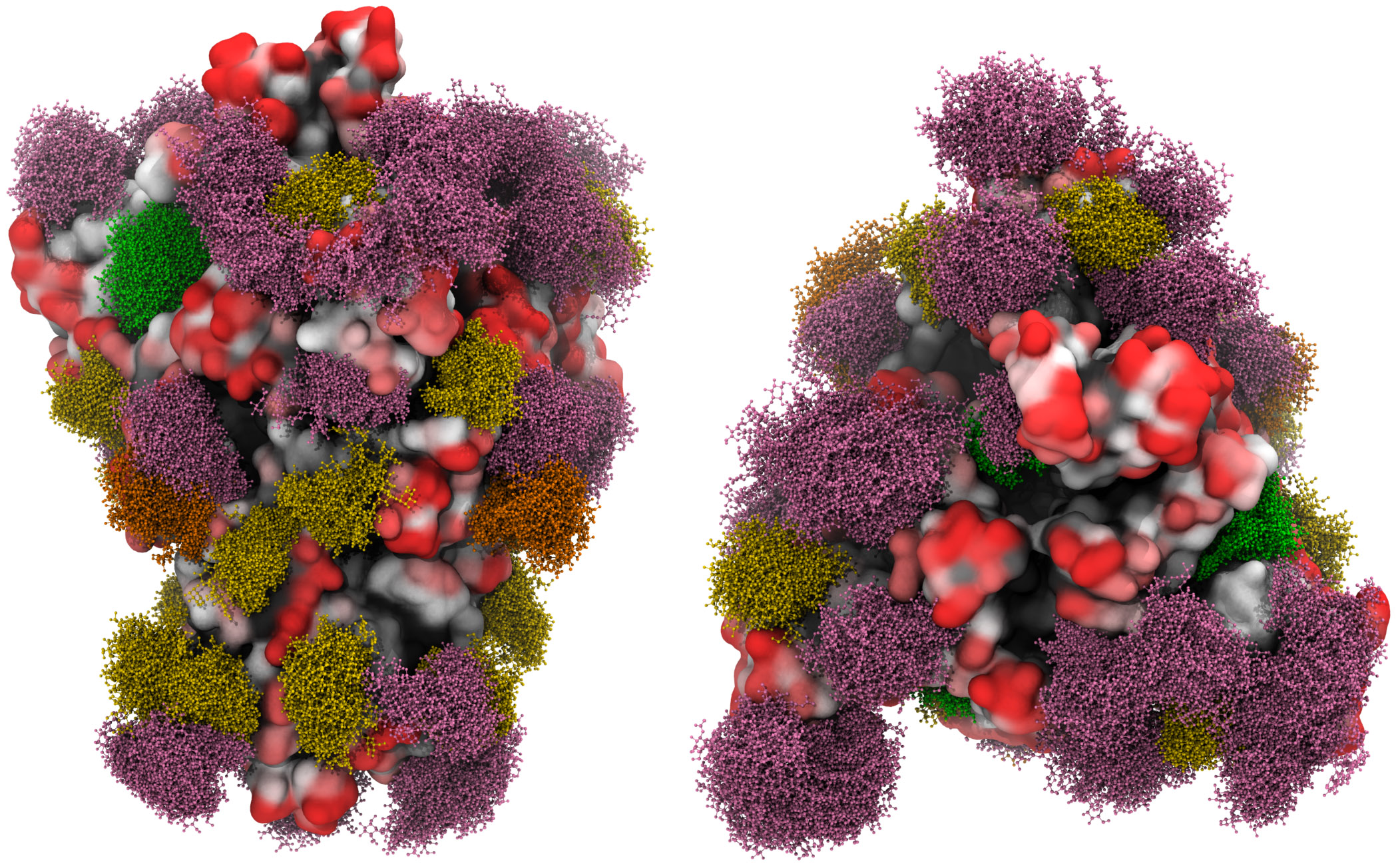
A three-dimensional molecular simulation of the spike glycoprotein with the moss effect showing the glycan shielding [Grant et al, 2020].
Even with an exoribonuclease, there will be errors in transcription and replication. These errors exist for a while and appear as new strains before disappearing. Certainly not a new mutation then.
Nucleic Acid Testing Is Error-Prone.
As we have written about previously, nucleic acid testing has many drawbacks and should not be relied upon as the basis for decision-making. The cycling and amplification of the RT-PCR process mean that it is prone to errors. In another previous article, we set out how RT-PCR nucleic aid testing works (or more accurately, doesn’t), explaining how it looks for tiny fragments of the viral RNA. It does not show when or where infection occurred, and neither does it show if the amino acid or peptide chains are alive or dead (and if so, how long ago they died).
The homology of beta genus coronavirus genomes, as we have set out here, means that using nucleic acid testing as a way of ‘testing for the coronavirus’ is incorrect. You are in fact testing for any coronavirus because the tiny fragments of viral RNA in embecovirus/lineage A betacoronaviruses are the same as those in sarbecovirus/lineage B betacoronaviruses. They share a familial relationship and identical peptide chains across elements of their respective genomes.
Nucleic Acid Testing Is Picking Up The Common Cold.
RT-PCR testing is like going through somone’s bins and finding a fragment of a chocolate bar wrapper with the letter ‘C’ written on it. In the government’s flawed strategy, this means testing positive for the Chocolate bar. No, it means testing positive for a Cadbury’s chocolate bar, of which there are at 83 (source: Cadbury). RT-PCR is incapable of detecting which one.
Using nucleic acid testing in identifying the number of viral mutations leads to overestimation of the mutation frequency [Sanjuán et al, 2010]. Applying this to the above example, you would have one correct result and 82 mutations.
As embecovirus/lineage A betacoronaviruses HCoV-HKU1 and HCOV-OC43 cause the common cold and share their genetic sequence with sarbecovirus/lineage B betacoronavirus SARS-CoV-2, RT-PCR nucleic acid testing can’t tell them apart.
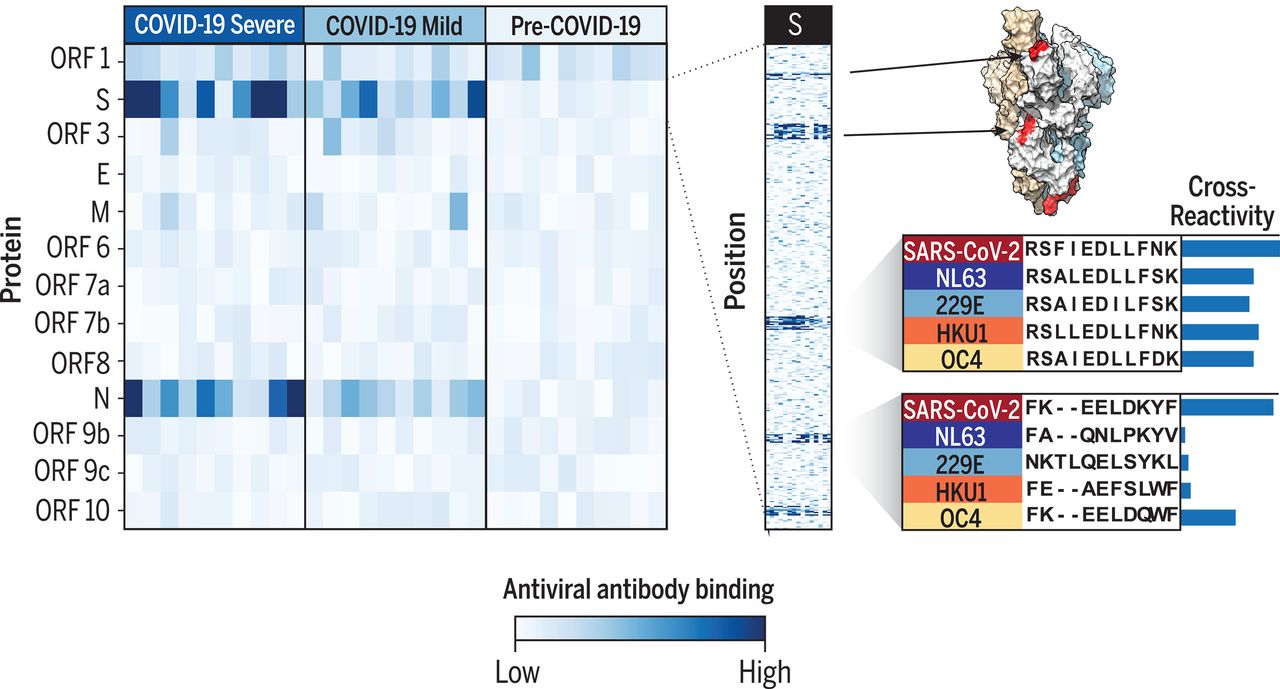
Cross-reactive epitopes of SARS-CoV-2 and the other four common-cold causing coronaviruses [Shrock et al, 2020].
So, a good proportion of these increasing number of positive tests are indeed testing positive…for the common cold.