The Four Nucleobases.
Almost exactly twelve months ago, we explained how increased transmissibility also meant greater vulnerability, i.e. a new variant may be more infectious but it is also more susceptible to detection by the immune system. This was first evidenced by our study of SARS-CoV-2-SG614, also one year ago now. Despite there being virtually no international travel during the period, G614 became the fixed, global dominant variant between March and June 2020. It did this as a result of one single nucleobase variation, adenine to guanine, at position 23403 in SARS-CoV-2’s genome, which caused the D614G amino acid substitution. What is of interest now are the other two nucleobases, cytosine and uracil.
G614 became 2020’s dominant variant, showing a 9-fold increase in infectiousness but no increase in infectivity [Korber et al., 2020; Groves et al., 2020; Zhang et al., 2020]. In 2021 there was another amino acid substitution, T478K, which became the lead amino acid substitution in the Delta variant (G614 occured before the current nomenclature was adopted). As with G614, T478K showed increased infectiousness but no increased infectivity, with the data starting to indicate reduced infectivity.
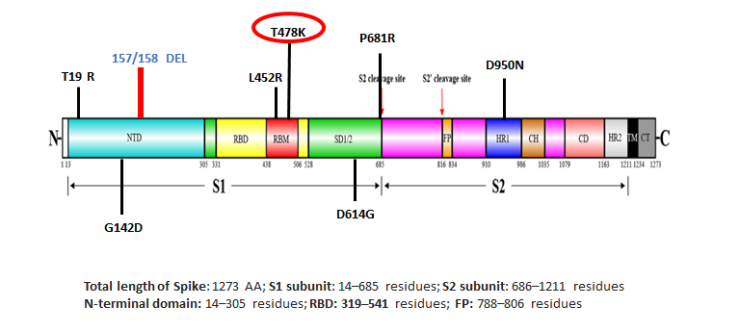
Delta variant structural spike protein domain, showing location of T478K. Note the presence of D614G. Source: https://science.thewire.in.
In late 2021 and into 2022 another dominant, global variant emerged, led by amino acid substitutions N440K and Q493R, designated Omicron. The key difference this time around is increased infectiousness but with substantially reduced infectivity, ranging from 40%-45% lower (source: Imperial College London), to 50%-70% lower (source: UK Health Security Agency) to 80% lower (Cohen 𝘦𝘵 𝘢𝘭.) This should not be surprising to anyone since viruses attenuate as they evolve, becoming weaker with each ‘wave’ or variant.
As early as September 2020 we had identified that SARS-CoV-2’s infectivity was attenuating, i.e. it was starting to weaken. This is in line with how viruses evolve phylogenetically. With each ‘wave’ or dominant variant, the infectivity reduces as a result principally of increased immunity, especially herd immunity. Remember that the majority of the United Kingdom population had been exposed to SARS-CoV-2 by 19th March 2020, with Oxford University’s Evolutionary Ecology of Infectious Disease study – undertaken on a near real-time basis during peak infections – showing an infection rate in the range 68%-80% depending upon the R0 ratio applied. Something we highlighted first in April 2020.
In addition to natural and to a degree antiviral-induced immunity (ignoring the impact of escape neutralisation variants on COVID-19 antivirals), another factor has been at play. It is a component of the innate immune response and it presents within every individual.
APOBEC-Mediated Cytosine To Uracil Editing.
It proves that variants are not geographical in nature and do not spread geographically. We first evaluated this in March 2021 Why Geographical Naming Of Variants Is Wrong and expanded upon it later that month The ‘Third Wave’ That Has Been Here All Along. The reason is cytosine to uracil editing (C-to-U editing) mediated by the apolipoprotein-B mRNA editing enzyme catalytic polypeptide-like protein (APOBEC) family of enzymes.
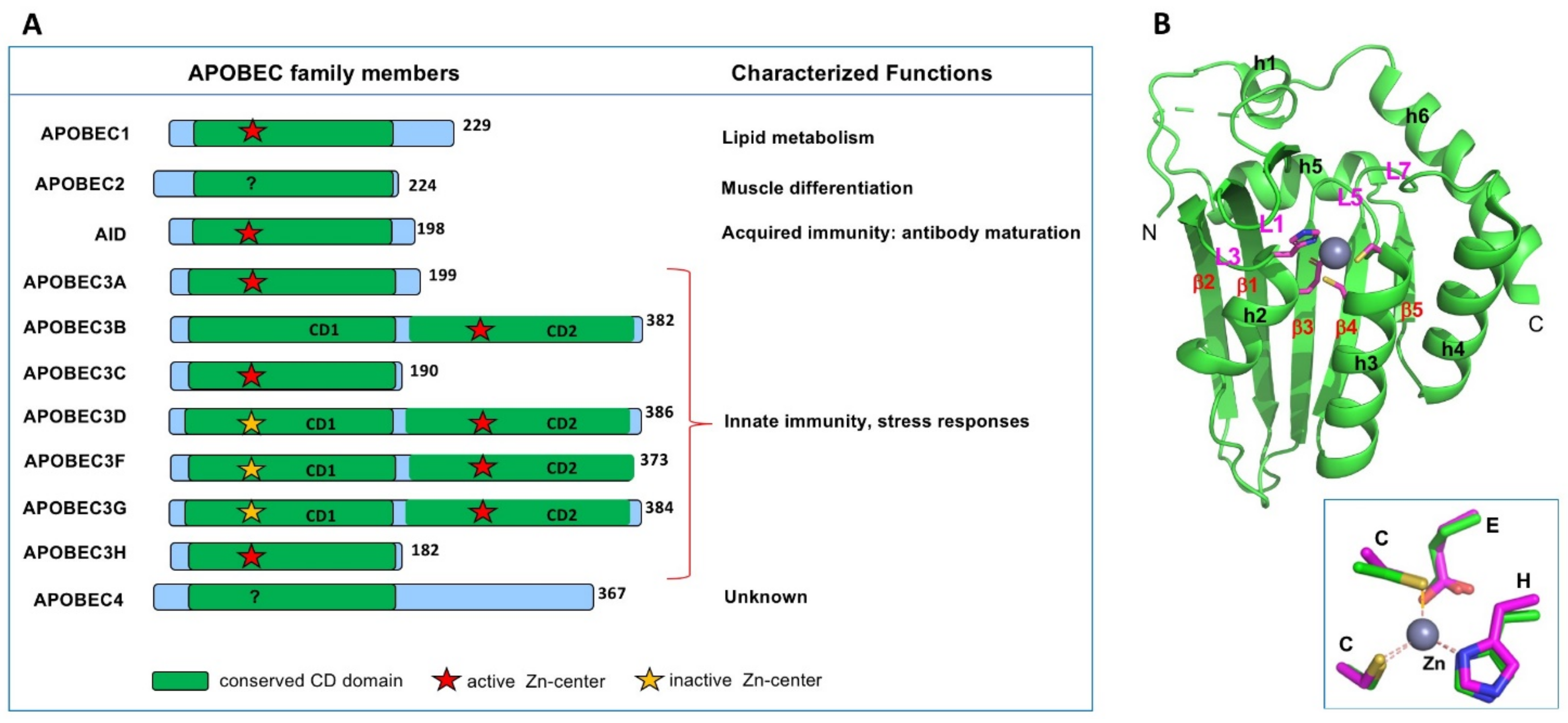
APOBEC family (A) and illustrative core structure of APOBEC A subfamily. Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions, Chen 2021; doi.org/10.3390/v13030497
APOBEC enzymes function on an intracellular basis and form part of the innate immune response where, upon detection of a virus, they inhibit its replication cycle. This is done through targeting nucleobases within individual nucleotides, deaminating cytosine to uracil. This editing interrupts the transcription of the mRNA by the RNA-dependent RNA polymerase (RdRp).
The coronaviridae family is distinct but not unique among viruses in containing an exoribonuclease. In SARS-CoV-2 it is contained in nonstructural protein 14 (nsp14-ExoN) and it functions like a proofreader, monitoring the replication cycle and correcting mistakes made by the RdRp. These mistakes can be random errors but can also be deliberate as part of natural selection, where the virus is constantly seeking to gain a fitness advantage, either through increased infectiousness (not infectivity) and/or immunoevasion. Think of it as quality control, overseeing the hijakced ribosome of the infected host cell as it makes lots of copies of the virus’ genome.
There is an inverse correlation between RdRp fidelity and recombination: the higher the fidelity, the lower the recombination. The presence of nsp14-ExoN thereby maintains high fidelity replication. However, C-to-U editing is very subtle as it is performed at the single nucleotide level, allowing it to go undetected by nsp14-ExoN.
Within the APOBEC family, C-to-U editing is a normal part of the innate immune response with APOBEC A1 [Driscoll 𝘦𝘵 𝘢𝘭., 1989; Teng 𝘦𝘵 𝘢𝘭.,1993], A3A and A3G [Sharma 𝘦𝘵 𝘢𝘭., 2015/2016] targeting single-stranded RNA viruses. Kim 𝘦𝘵 𝘢𝘭. [2021] show that across all single nucleotide variations in the entire SARS-CoV-2 genome, C-to-U accounts for approx. 54%, making it the dominant change by a significant margin.
This is important as if you change a nucleotide you can change a codon; change a codon and you can change an amino acid. As above, SARS-CoV-2 variants are all about amino acid substitutions. It also means that C-to-U editing is one of the main triggers of SARS-CoV-2 variants.
Variants Are Triggered By C-To-U Editing Within The Innate Immune Response.
They are triggered automatically by the virus:host interaction and are part of the innate immune response. C-to-U editing degrades the virus’ genome during its replication cycle. It is nothing to do with geography or travel. This is why D614G became the dominant, global variant from March-June 2020 when there was hardly any international travel. Variants have occurred and will continue to occur as long as the virus is in existence and regardless of any travel restrictions or attempts to control travel.
As with most biological actions, there is a function and counterfunction. On the one side, SARS-CoV-2 exploits APOBEC in an attempt to gain fitness advantage that will not be detected by the exoribonuclease. On the other, with each new, dominant ‘wave’ or variant that emerges, APOBEC-mediated editing ensures that the fidelity of replication has been degraded. Therefore, while APOBEC’s usual function as an effective antiviral countermeasure exacerbates the number of variants that will exist, it ensures that those variants become weaker.
You, me, all of us are the causes of SARS-CoV-2 variants but as part of the immune system’s progressive weakening of those variants.